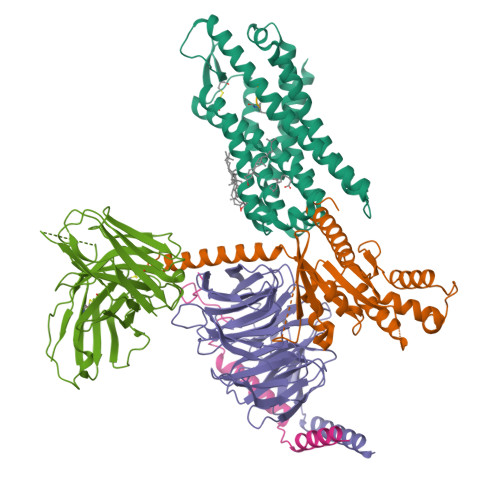
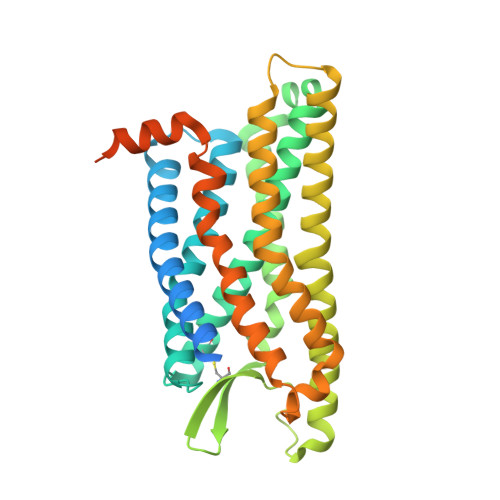
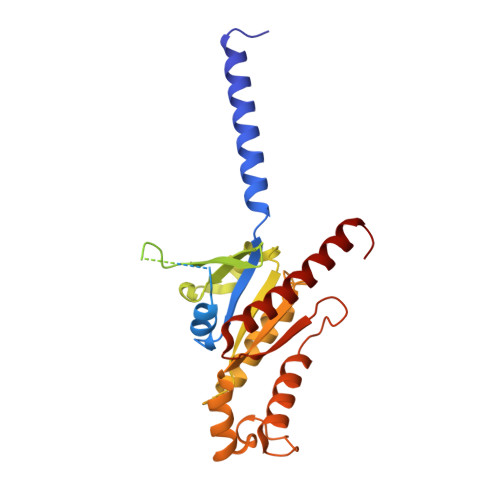
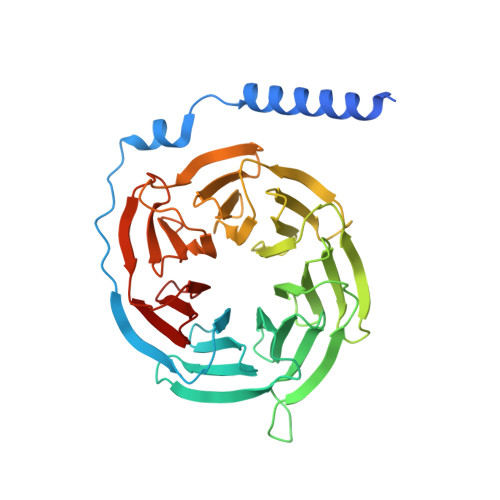
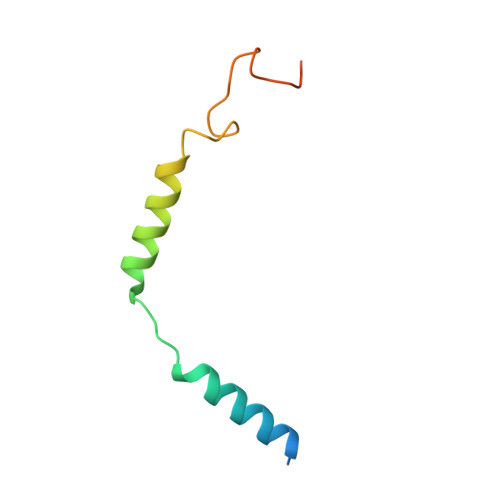
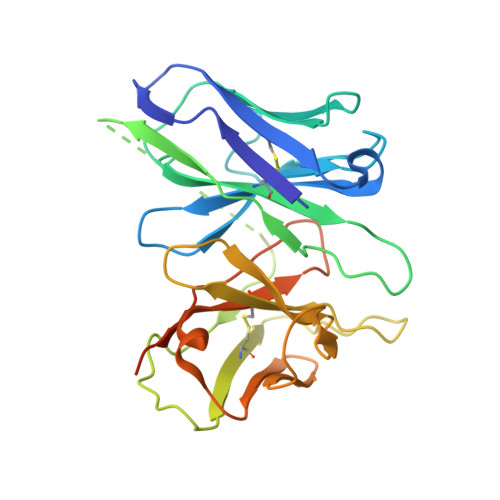
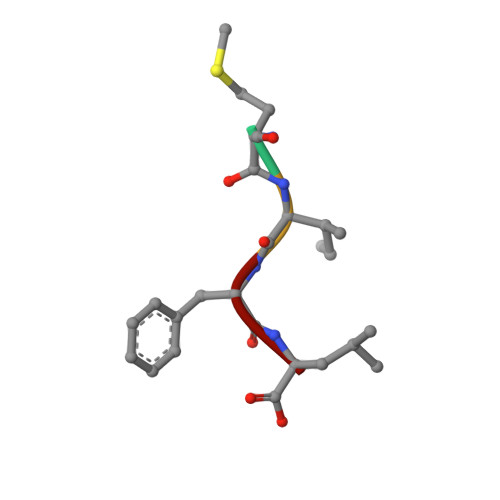
Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Chen, G., Wang, X., Liao, Q., Ge, Y., Jiao, H., Chen, Q., Liu, Y., Lyu, W., Zhu, L., van Zundert, G.C.P., Robertson, M.J., Skiniotis, G., Du, Y., Hu, H., Ye, R.D.(2022) Nat Commun 13: 5232-5232
- PubMed: 36064945
- DOI: https://doi.org/10.1038/s41467-022-32822-y
- Primary Citation of Related Structures:
7EUO, 7VFX - PubMed Abstract:
The formyl peptide receptor 1 (FPR1) is primarily responsible for detection of short peptides bearing N-formylated methionine (fMet) that are characteristic of protein synthesis in bacteria and mitochondria. As a result, FPR1 is critical to phagocyte migration and activation in bacterial infection, tissue injury and inflammation. How FPR1 distinguishes between formyl peptides and non-formyl peptides remains elusive. Here we report cryo-EM structures of human FPR1-Gi protein complex bound to S. aureus-derived peptide fMet-Ile-Phe-Leu (fMIFL) and E. coli-derived peptide fMet-Leu-Phe (fMLF). Both structures of FPR1 adopt an active conformation and exhibit a binding pocket containing the R201 5.38 XXXR205 5.42 (RGIIR) motif for formyl group interaction and receptor activation. This motif works together with D106 3.33 for hydrogen bond formation with the N-formyl group and with fMet, a model supported by MD simulation and functional assays of mutant receptors with key residues for recognition substituted by alanine. The cryo-EM model of agonist-bound FPR1 provides a structural basis for recognition of bacteria-derived chemotactic peptides with potential applications in developing FPR1-targeting agents.
Organizational Affiliation:
Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong, 518172, China.